Introduction to skull stripping (Image segmentation on 3D MRI images)
This article was published as a part of the Data Science Blogathon
Skull stripping is one of the preliminary steps in the path of detecting abnormalities in the brain. It is the process of isolating brain tissue from non-brain tissue from an MRI image of a brain. This segmentation of the brain from the skull is a tedious task even for expert radiologists and the accuracy of results varies a lot from person to person. Here we are trying to automate the process by creating an end-to-end pipeline where we just need to input the raw MRI image and the pipeline should output a segmented image of the brain after doing the necessary preprocessing.
So what is an MRI image?
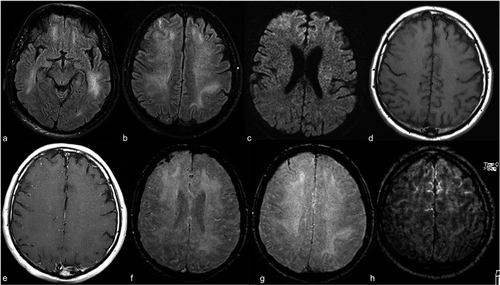
To get an MR image of a patient they are inserted into a tunnel with a magnetic field inside. This causes all protons in the body to ‘align’ themselves so their quantum spin is the same. A pulse of the oscillating magnetic field is then used to disrupt this alignment. When the protons return to equilibrium they send out an electromagnetic wave. Based on fat content, chemical composition, importantly type of stimulation (i.e. sequences) used to disrupt the protons, different images will be obtained. Four common sequences that are obtained are T1, T1 with contrast (T1C), T2, and FLAIR.
Common challenges while working with brain images
-
Lack of large datasets like Imagenet or Coco.
Generating gold standard data is a very time-consuming task and needs to be done by experts. Most available data sets for skull stripping are very small in size in order to build a deep model.
-
Domain-specific knowledge required for preprocessing
Before feeding the image to the model several preprocessing steps need to be done on the image. This requires domain-specific knowledge.
-
Challenges on real-world data
Building a model and achieving a good accuracy on a jupyter notebook is nice. But most of the time a very good performing model performs very badly on real-world data. This happens due to data drift when the model sees completely different data than what it is trained on. In our case, it can happen due to differences in some parameters or methods of generating MRI images. Here is a blog outlining some failures of AI in the real world.
Problem formulation
The task we have here is to give a 3D MRI image we have to identify the brain and segment the brain tissue from the entire image of a skull. For this task, we will be having a ground truth label and hence it will be a supervised image segmentation task. We will be using dice loss as our loss function.
Data
Let’s have a look at the data set we will be using for this task. The data set can be downloaded from here.
The repository contains data from 125 participants, 21 to 45 years old, with a variety of clinical and sub-clinical psychiatric symptoms. For each participant, the repository contains:
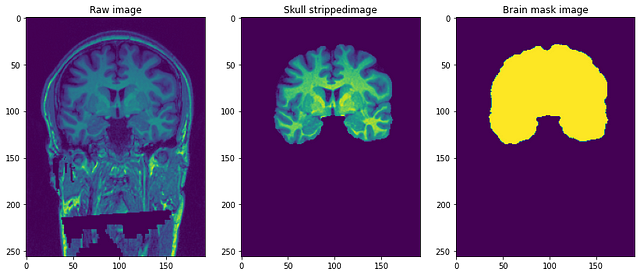
- Structural T1-weighted anonymized (de-faced) image: This is the raw T1weighted MRI image with a single channel.
- Brain mask: It is the image mask of the brain or can be called the ground truth. It is obtained using the Beast(Brain extraction based on nonlocal segmentation) method and applying manual edits by domain experts to remove non-brain tissue.
- Skull-stripped image: This can be thought of as part of the brain stripped from the above T1weighted image. This is similar to overlaying masks to actual images.
The resolution of the images is 1 mm3 and each file is in NiFTI format (.nii.gz). A single data point looks something like this..
Preprocessing our Raw images
img=nib.load('/content/NFBS_Dataset/A00028185/sub-A00028185_ses-NFB3_T1w.nii.gz')
print('Shape of image=',img.shape)

Imagine above 3-D images like we have 192 2-D images of size 256*256 stacked on top of each other.
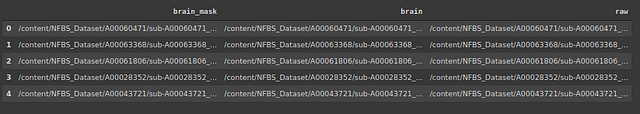
Let’s create a data frame containing the location of images and corresponding masks and skull-stripped images.
#storing the address of 3 types of files
import os
brain_mask=[]
brain=[]
raw=[]
for subdir, dirs, files in os.walk('/content/NFBS_Dataset'):
for file in files:
#print os.path.join(subdir, file)y
filepath = subdir + os.sep + file
if filepath.endswith(".gz"):
if '_brainmask.' in filepath:
brain_mask.append(filepath)
elif '_brain.' in filepath:
brain.append(filepath)
else:
raw.append(filepath)
-
Bias field correction
Bias field signal is a low-frequency and very smooth signal that corrupts MRI images especially those produced by old MRI (Magnetic Resonance Imaging) machines. Image processing algorithms such as segmentation, texture analysis, or classification that use the gray level values of image pixels will not produce satisfactory results. A pre-processing step is needed to correct for the bias field signal before submitting corrupted MRI images to such algorithms or the algorithms should be modified.
-
Cropping and Resizing
Due to computational limitations of fitting complete image to model here, we decide to reduce the size of the MRI image from (256*256*192) to (96*128*160). The target size is chosen in such a way that most part of the skull is captured and after cropping and resizing it has a centering effect on the images.
-
Intensity normalization
Normalization shifts and scales an image so that the pixels in the image have a zero mean and unit variance. This helps the model to converge faster by removing scale in-variance. Below is the code for it.
class preprocessing(): def __init__(self,df): self.data=df self.raw_index=[] self.mask_index=[] def bias_correction(self): !mkdir bias_correction n4 = N4BiasFieldCorrection() n4.inputs.dimension = 3 n4.inputs.shrink_factor = 3 n4.inputs.n_iterations = [20, 10, 10, 5] index_corr=[] for i in tqdm(range(len(self.data))): n4.inputs.input_image = self.data.raw.iloc[i] n4.inputs.output_image ='bias_correction/'+str(i)+'.nii.gz' index_corr.append('bias_correction/'+str(i)+'.nii.gz') res = n4.run() index_corr=['bias_correction/'+str(i)+'.nii.gz' for i in range(125)] data['bias_corr']=index_corr print('Bias corrected images stored at : bias_correction/') def resize_crop(self): #Reducing the size of image due to memory constraints !mkdir resized target_shape = np.array((96,128,160)) #reducing size of image from 256*256*192 to 96*128*160 new_resolution = [2,]*3 new_affine = np.zeros((4,4)) new_affine[:3,:3] = np.diag(new_resolution) # putting point 0,0,0 in the middle of the new volume - this could be refined in the future new_affine[:3,3] = target_shape*new_resolution/2.*-1 new_affine[3,3] = 1. raw_index=[] mask_index=[] #resizing both image and mask and storing in folder for i in range(len(data)): downsampled_and_cropped_nii = resample_img(self.data.bias_corr.iloc[i], target_affine=new_affine, target_shape=target_shape, interpolation='nearest') downsampled_and_cropped_nii.to_filename('resized/raw'+str(i)+'.nii.gz') self.raw_index.append('resized/raw'+str(i)+'.nii.gz') downsampled_and_cropped_nii = resample_img(self.data.brain_mask.iloc[i], target_affine=new_affine, target_shape=target_shape, interpolation='nearest') downsampled_and_cropped_nii.to_filename('resized/mask'+str(i)+'.nii.gz') self.mask_index.append('resized/mask'+str(i)+'.nii.gz') return self.raw_index,self.mask_index def intensity_normalization(self): for i in self.raw_index: image = sitk.ReadImage(i) resacleFilter = sitk.RescaleIntensityImageFilter() resacleFilter.SetOutputMaximum(255) resacleFilter.SetOutputMinimum(0) image = resacleFilter.Execute(image) sitk.WriteImage(image,i) print('Normalization done. Images stored at: resized/')
Let’s have a look at the architecture of the model.
def data_gen(self,img_list, mask_list, batch_size):
'''Custom data generator to feed image to model'''
c = 0
n = [i for i in range(len(img_list))] #List of training images
random.shuffle(n)
while (True):
img = np.zeros((batch_size, 96, 128, 160,1)).astype('float') #adding extra dimensions as conv3d takes file of size 5
mask = np.zeros((batch_size, 96, 128, 160,1)).astype('float')
for i in range(c, c+batch_size):
train_img = nib.load(img_list[n[i]]).get_data()
train_img=np.expand_dims(train_img,-1)
train_mask = nib.load(mask_list[n[i]]).get_data()
train_mask=np.expand_dims(train_mask,-1)
img[i-c]=train_img
mask[i-c] = train_mask
c+=batch_size
if(c+batch_size>=len(img_list)):
c=0
random.shuffle(n)
yield img,mask
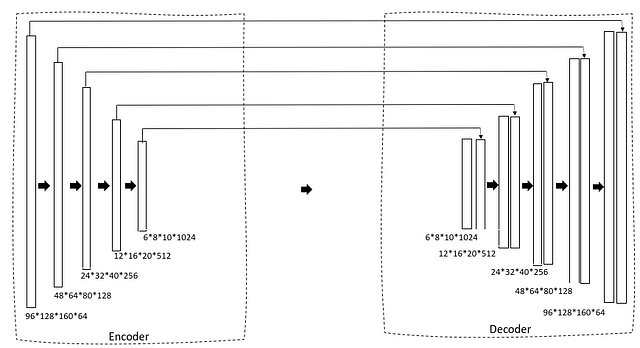
We are using a 3D U-Net as our architecture. If you are already familiar with the 2D U-Net this is going to be very simple. First, we have a contracting path via an encoder that gradually reduces the size of the image and the number of filters is increased to generate bottleneck features. This is then fed into a decoder block that gradually expands the size so that finally it can generate a mask as predicted output.
def convolutional_block(input, filters=3, kernel_size=3, batchnorm = True):
'''conv layer followed by batchnormalization'''
x = Conv3D(filters = filters, kernel_size = (kernel_size, kernel_size,kernel_size),
kernel_initializer = 'he_normal', padding = 'same')(input)
if batchnorm:
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = Conv3D(filters = filters, kernel_size = (kernel_size, kernel_size,kernel_size),
kernel_initializer = 'he_normal', padding = 'same')(input)
if batchnorm:
x = BatchNormalization()(x)
x = Activation('relu')(x)
return x
def resunet_opt(input_img, filters = 64, dropout = 0.2, batchnorm = True):
"""Residual 3D Unet"""
conv1 = convolutional_block(input_img, filters * 1, kernel_size = 3, batchnorm = batchnorm)
pool1 = MaxPooling3D((2, 2, 2))(conv1)
drop1 = Dropout(dropout)(pool1)
conv2 = convolutional_block(drop1, filters * 2, kernel_size = 3, batchnorm = batchnorm)
pool2 = MaxPooling3D((2, 2, 2))(conv2)
drop2 = Dropout(dropout)(pool2)
conv3 = convolutional_block(drop2, filters * 4, kernel_size = 3, batchnorm = batchnorm)
pool3 = MaxPooling3D((2, 2, 2))(conv3)
drop3 = Dropout(dropout)(pool3)
conv4 = convolutional_block(drop3, filters * 8, kernel_size = 3, batchnorm = batchnorm)
pool4 = MaxPooling3D((2, 2, 2))(conv4)
drop4 = Dropout(dropout)(pool4)
conv5 = convolutional_block(drop4, filters = filters * 16, kernel_size = 3, batchnorm = batchnorm)
conv5 = convolutional_block(conv5, filters = filters * 16, kernel_size = 3, batchnorm = batchnorm)
ups6 = Conv3DTranspose(filters * 8, (3, 3, 3), strides = (2, 2, 2), padding = 'same',activation='relu',kernel_initializer='he_normal')(conv5)
ups6 = concatenate([ups6, conv4])
ups6 = Dropout(dropout)(ups6)
conv6 = convolutional_block(ups6, filters * 8, kernel_size = 3, batchnorm = batchnorm)
ups7 = Conv3DTranspose(filters * 4, (3, 3, 3), strides = (2, 2, 2), padding = 'same',activation='relu',kernel_initializer='he_normal')(conv6)
ups7 = concatenate([ups7, conv3])
ups7 = Dropout(dropout)(ups7)
conv7 = convolutional_block(ups7, filters * 4, kernel_size = 3, batchnorm = batchnorm)
ups8 = Conv3DTranspose(filters * 2, (3, 3, 3), strides = (2, 2, 2), padding = 'same',activation='relu',kernel_initializer='he_normal')(conv7)
ups8 = concatenate([ups8, conv2])
ups8 = Dropout(dropout)(ups8)
conv8 = convolutional_block(ups8, filters * 2, kernel_size = 3, batchnorm = batchnorm)
ups9 = Conv3DTranspose(filters * 1, (3, 3, 3), strides = (2, 2, 2), padding = 'same',activation='relu',kernel_initializer='he_normal')(conv8)
ups9 = concatenate([ups9, conv1])
ups9 = Dropout(dropout)(ups9)
conv9 = convolutional_block(ups9, filters * 1, kernel_size = 3, batchnorm = batchnorm)
outputs = Conv3D(1, (1, 1, 2), activation='sigmoid',padding='same')(conv9)
model = Model(inputs=[input_img], outputs=[outputs])
return model
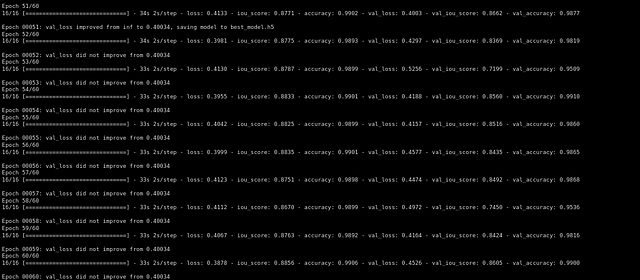
Then we trained the model using Adam optimizer and dice loss as our loss function…
def training(self,epochs):
im_height=96
im_width=128
img_depth=160
epochs=60
train_gen = data_gen(self.X_train,self.y_train, batch_size = 4)
val_gen = data_gen(self.X_test,self.y_test, batch_size = 4)
channels=1
input_img = Input((im_height, im_width,img_depth,channels), name='img')
self.model = resunet_opt(input_img, filters=16, dropout=0.05, batchnorm=True)
self.model.summary()
self.model.compile(optimizer=Adam(lr=1e-1),loss=focal_loss,metrics=[iou_score,'accuracy'])
#fitting the model
callbacks=callbacks = [
ModelCheckpoint('best_model.h5', verbose=1, save_best_only=True, save_weights_only=False)]
result=self.model.fit(train_gen,steps_per_epoch=16,epochs=epochs,validation_data=val_gen,validation_steps=16,initial_epoch=0,callbacks=callbacks)
After training for 60 epochs we got a validation iou_score of 0.86.
Let’s have a look at how our model performed. Our model will be predicting simply the mask. To get the skull stripped image we need to overlay it on the Raw image to get skull stripped image…
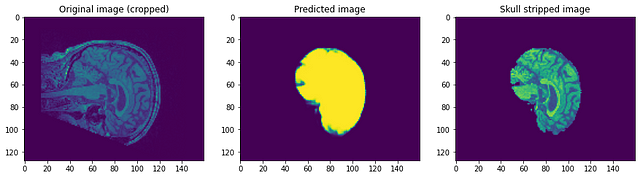
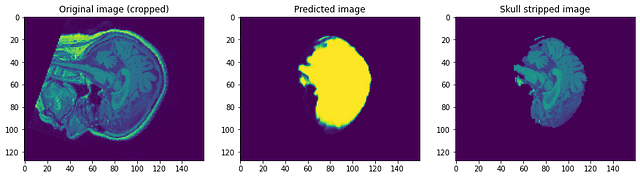

Looking at the predictions we can say that although it is able to identify the brain and segment it, it is nowhere near perfection. At this point, we can sit with a domain expert to identify what further preprocessing steps can be done to improve accuracy. But as for this post, I will conclude it here. Please follow link1 and/or link2 if you want to know more…
Conclusion:
Glad you made it to the end.Hope this helps you in getting started with image segmentation on 3D images. You can find the google colab link containing the code here. Please feel free to add any suggestions or queries in the comment section. Have a nice day!
The media shown in this article on skull stripping are not owned by Analytics Vidhya and are used at the Author’s discretion.









Hey hello Bai Dash Your blog/ article is good to follow for sequence only.. But there are some mistakes which I feel they are (maybe may not) def convo_block: => have some mistake or is it intentionally repeated con3D commands..?????? N4Biasfield correction also not working properly ...some issue with lib and code The data generator also has an issue... and many more... Kindly provide the better version which might have attentions of 100x views