Personalized Medicine through Machine Learning
This article was published as a part of the Data Science Blogathon
Introduction
Medicine is the key to maintain and prolong life. Not all body systems are clinically similar, so the medicine needs to be customized according to the body system of an individual. The ongoing pandemic acts as a burning example as it has been observed that one the set of medicines like Remdisivir, Tocilizumab, etc. works for one category of patients while the same set of medicines cannot prevent another category of patients with almost similar clinical parameters from progressing to severe stage from mild or moderate condition. Personalized medicine can be a solution to this challenge as it has a more “customized” approach. It is also known as precision/individualized/customized medicine
Machine Learning (ML) is often pooled with Artificial Intelligence (AI). However, ML is a branch of AI that identifies variable patterns of data to predict or classify hidden or unseen patterns which in turn can be used for exploratory data analysis, data mining, and data modeling. The ML algorithms indicate the possibility of identifying target-based medicines based upon clinical, genomics, laboratory, nutrition, and lifestyle-related data.
Throughout this article acronyms like ML, and AI, and terminology personalized medicine would be used interchangeably with Machine Learning, and Artificial Intelligence, and precision/individualized/customized medicine. It is assumed that the readers of this article have basic knowledge of medical terminologies, python, and data science.
Image Source: KDnuggets
Benefits of personalized medicine
1. It would reduce trial and error-based treatment decisions.
2. It would bring down the burdens associated with a condition both in terms of health and finance.
3. Patient-centric medication through the integration of multi-modal data from an individual.
4. More emphasis would be laid on preventive mode rather than the reactive mode in medicine
5. Reduction in time, and cost associated with clinical trials conducted by pharmaceuticals.
Role of Machine Learning
1. There is a scope of applying the algorithms of machine learning to the genomic datasets which would enable the delivery of personalized medicines.
2. The use of multi-modal data helps in deeper analysis of large datasets which improves the understanding of human health and disease by leaps and bound.
3. As ML is capable of identifying hidden patterns of data, many future diseases can be prevented.
4. Advancement in the field of “in silico” experimental systems would improve the efficiency of clinical trials which would reduce the time and cost associated with clinical trials. The experimental system “in silico” refers to using computers to run various experiments (Wanner, 2021).
5. Reduction of the burden on the healthcare system on screening of various diseases of seriousness like lung cancer, covid19, heart diseases, etc.
Challenges for Machine Learning in the field of Personalized Medicine
1. Optimization of application is required.
2. The knowledge base of the stakeholders which would involve physicians, laboratory technicians, data analysts, programmers, and paramedical staff has to be increased. Everyone needs to have a basic understanding of the concerned domains.
Tools of Machine Learning for Precision Medicine
There are three primary tools for machine learning algorithms. These are classification, regression, and clustering. Let’s have a look at the basic concept of each of these.
1. Classification – Logistic Regression and Naive Bayes are the most common supervised learning classification algorithms.
2. Regression – Linear Regression is the most common supervised learning regression algorithm.
3. Clustering – K-means Algorithm, Mean Shift Algorithm, and Hierarchical Clustering are the common algorithms. These are all unsupervised, i.e. target variable is not available.
4. Classification and Regression combined – Support Vector Machine (SVM), Decision Tree, Random Forest, and K-Nearest Neighbors are types of supervised ML algorithms that are applicable in both classification and regression predictive problems.
Another very important ML type is Reinforcement Learning which is applied when a categorical target variable is available as well as when no target variable is available. It has a got wide application in the area of auto-car and optimized marketing. It is a semi-supervised algorithm.
In this article, we would restrict ourselves to a few ML algorithms which are exclusively used in precision medicine.
Machine Learning and Precision Medicine in real world
The purpose of personalized medicine is to select and deliver patient-specific treatments to achieve the best possible outcome. The challenge lies in identifying an optimum treatment as the number of possible predictors of good response like genetic and other biomarkers, and the option of treatments is increasing.
In addition to this, as most clinical trials are based upon average treatment effects, similar medicines become non-responsive for some patients and responsive for some other patients.
An example in this regard is the primary analysis of the COMBINE Study which is one of the largest clinical trials regarding treatments for alcohol dependence in the USA. The study inferred that there was an impact of one of the considered pharmacological treatments (naltrexone) but was non-responsive for another, acamprosate (Tsai et al., 2016).
CART (Classification and Regression Trees) methods consider a large number of potential predictors and identify combinations of patient characteristics and good outcomes. Personalized medicine has a focus on whom a particular treatment may be more effective than that of another. The application of a modified tree-based approach indicates the possibility of selection of the best individualized treatment based on baseline features (Tsai et al., 2016).
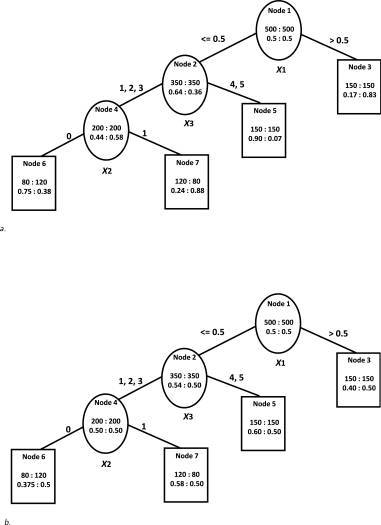
Image Source: Tsai et al., 2016
The approach of modern-day medicine is based upon a population-wide model which is intended to be applied to the overall population and is optimized to have decent predictive performance on an average number of people out of that population. This approach has done remarkably well for decades but it ignores individual differences in treatment responses.
A better approach for capturing individual differences in treatment responses is the patient-specific modeling approach. The personalized decision tree model is a patient-specific modeling approach that performed a bit better than the CART method (Adam, & Aliferis, 2019).
Gini impurity, information entropy, and variance reduction are 3 important metrics for decision tree algorithm. Gini impurity is the more preferred metric among the 3 metrics. It measures how a randomly chosen element is incorrectly labeled.
Criteria for constructing a tree
1. Every parent node of higher Gini impurity or information entropy is split into child nodes to lower its Gini impurity or information entropy.
Gini impurity of pure sets = 0
2. The preferred split between the 2 child nodes would be the one in which Gini impurity is higher would be split further.
3. Depending upon the complexity of parameters, the exploration of nodes is discontinued.
Packages and tools for precision medicine
1. Scikit-learn –
One of the most important tools for ML. It is an open-source library that aids in both supervised as well as unsupervised learning. From Scikit-learn various estimators or predictors are imported to model a particular dataset. Scikit provides us numerous models and ML algorithms. A few of them are
from sklearn.ensemble import RandomForestClassifier from sklearn.linear_model import LogisticRegression from sklearn.tree import DecisionTreeClassifier from sklearn.model_selection import train_test_split from sklearn.metrics import accuracy_score, confusion_matrix
2. pyGeno –
It is a Python package that pertains to precision medicine applications with a special focus on genomics and proteomics. It is easy to use, has highly efficient memory, and has a fast framework that allows users to easily explore subject-specific genomes and proteomes. pyGeno has been developed by creating a Python module that fully integrates within the Python environment making it user-friendly. The users can make use of functionalities like SciPy, NumPy 11, pandas, and matplotlib 13. It is available on https://pypi.python.org and can be installed by writing a simple command “pip install pyGeno”. Through get() function only, almost any query can be addressed and help() function helps in retrieving integrated documents (Daouda, Perreault, & Lemieuxb, 2016). Its last version was released on 29th February 2020.
pip install pyGeno
from pyGeno.Genome import * print(Exon.help())
Now, to build a personalized genome, we need to select the type of data we are interested in from the BioMart database. After selecting a database, we have to select a dataset that would be followed by filtration of the query. Then, we need to select BioMart attributes, by default it would be “Ensembl Gene ID” and “Ensembl Transcript ID”. Finally, the query will be displayed and retrieved. The details have been illustrated in the image below
import pyGeno.bootstrap as B B.printRemoteDatawraps()
A snapshot of the same has been provided below. It is to be noted that the file type is GZ so it has to be extracted with “Archive extractor online” or any other good extraction tools.
from pyGeno.Genome import * g = Genome(name = "GRCh37.75") prot = g.get(Protein, id = 'ENSP00000438917')[5] print (prot.sequence) print (prot.gene.biotype)
In the above lines, we are trying to extract protein sequence and gene biotype which would act as a reference set and would be used to create a personalized genome.
dummy = Genome(name = 'GRCh37.75', SNPs = 'dummySRY') dummy = Genome(name = 'GRCh37.75', SNPs = 'dummySRY', SNPFilter = myFilter()) dummy = Genome(name = 'GRCh37.75', SNPs = ['dummySRY', 'anotherSet'], SNPFilter = myFilter())
Above are steps for creating a personalized genome. It allows clinicians to work on the genomes and proteomes of patients. The entire working mechanism of pyGeno in the field of precision medicine can be seen in the image below.
Image Source: Daouda, Perreault, & Lemieuxb, 2016
Machine Learning, Precision Medicine, and ongoing Pandemic
In the ongoing pandemic, deciding upon the proper line of treatment for clinicians has become an enormous challenge. The clinicians are confused about the efficacy of remdisivir and corticosteroid on covid19 patients. ML algorithm can make a breakthrough in this area.
Lam et al. (2021) put forth that to evaluate the performance of corticosteroid versus remdesivir on identifying patients with longer survival times, Gradient-boosted decision-tree models were used. The models were trained and tested on data from 10 hospitals in the US on COVID-19 adult patients (age ≥18 years). Significant findings in treated and nontreated patients were based upon Fine and Gray proportional-hazards models.
The sample size was 2364 where 893 patients were treated with remdesivir, and 1471 were treated with a corticosteroid. The confounding was adjusted and it was found that in the populations identified by the algorithms, both corticosteroids and remdesivir were significantly associated with an increase in survival time, with hazard ratios of 0.56 and 0.40, respectively (both, P = 0.04). This contradicted the finding that neither corticosteroids nor remdesivir use were associated with increased survival time (Lam et al., 2021). This indicates that the ML algorithm holds promise in this field.
Conclusion
ML algorithms can identify at present which set of Covid19 patients would require Remdisivir and which set of patients would require corticosteroid so that the patient health outcome is improved. The algorithms can further be expanded into other areas of medicine as well. Nowadays, big biomedical data are in abundance, the need of the hour is to leverage these data for research in the field of medicine, public health, and biomedical procedures so that more and more people can be cured of serious diseases.
Thank You for your valuable time!
References
1. Aliferis, C., & Adams, T. (2019). Personalized and Precision Medicine Informatics. New York: Springer International Publishing.
2. Daouda, T., Perreault, C., & Lemieuxb, S. (2016). pyGeno: A Python package for precision medicine and proteogenomics. PMC. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5022704/
3. https://www.tutorialspoint.com/machine_learning_with_python/machine_learning_with_python_basics.htm
4. https://www.kdnuggets.com/
5. Lam, C et al. (2021). Machine Learning as a Precision-Medicine Approach to Prescribing COVID-19 Pharmacotherapy with Remdesivir or Corticosteroids. Clinical Therapeutics. Retrieved from https://www.sciencedirect.com/journal/clinical-therapeutics
6. Tsai, W. M., Zhang, H., Buta, E., O’Malley, S., & Gueorguieva, R. (2016). A modified classification tree method for personalized medical decisions. Statistics and its interface, 9(2), 239–253. Retrieved from https://doi.org/10.4310/SII.2016.v9.n2.a11
7. Wanner, M. (2021). MACHINE LEARNING: HOW DO YOU TEACH MACHINES.Tech Corner. The Jackson Laboratory. Retrieved from https://www.jax.org/news-and-insights/2021/february/machine-learning-how-do-you-teach-machines
The media shown in this article are not owned by Analytics Vidhya and are used at the Author’s discretion.









